Job Dispatcher Homepage
The Job Dispatcher frontpage can be accessed from here.
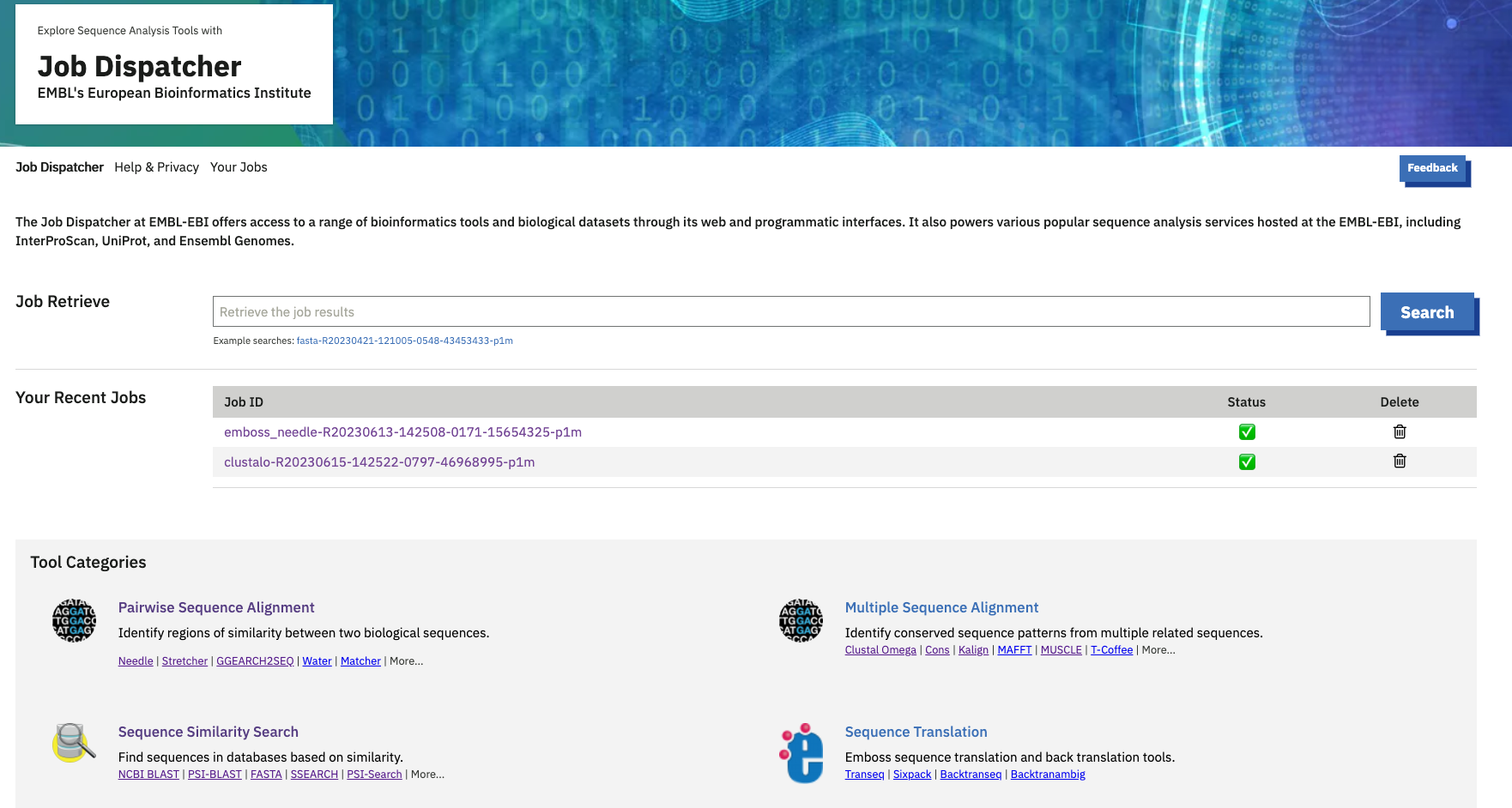
At the top, you can see the ‘Job Retrieve’ option to search for a job you already submitted. Provide your job ID and then search for it- this will take you to the respective tool result page. Multiple job searches cannot be possible from this search.
To view your recent job results, go to the "Your Recent Jobs" section. Here, you can see your five most recent jobs. If you want to see all your jobs, click ‘More’. Please note that job results are stored on our servers for seven days. It is recommended that you download the results for future reference.
You can also check the status of your job from the home page. If the job is completed without errors, the status will show as ‘Success’. If the job is not completed, the status will be ‘Failed. If the job ID is incorrect or unavailable, the status will display as ‘Not Found.
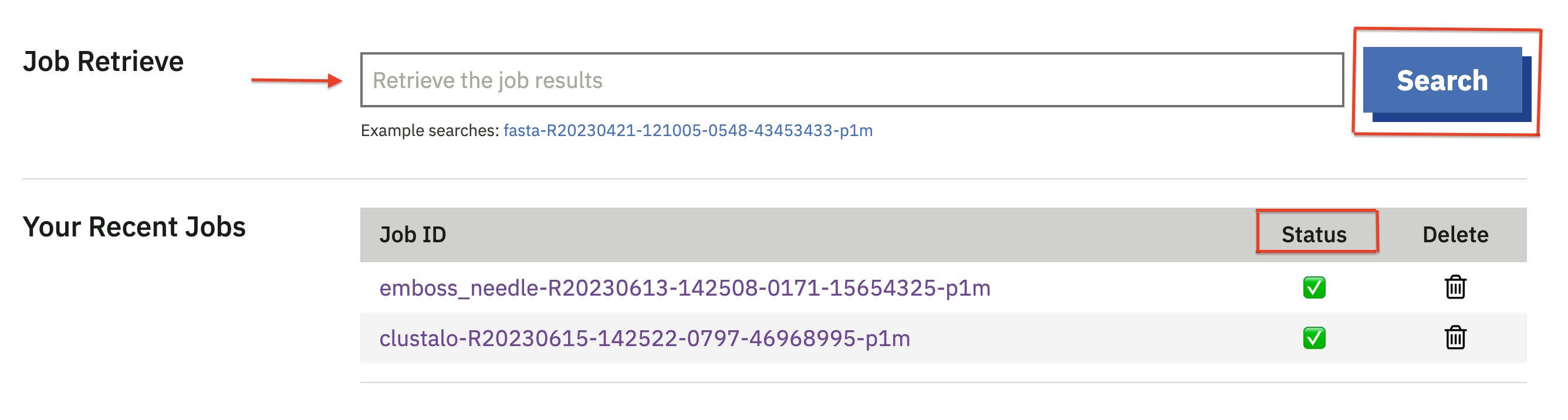
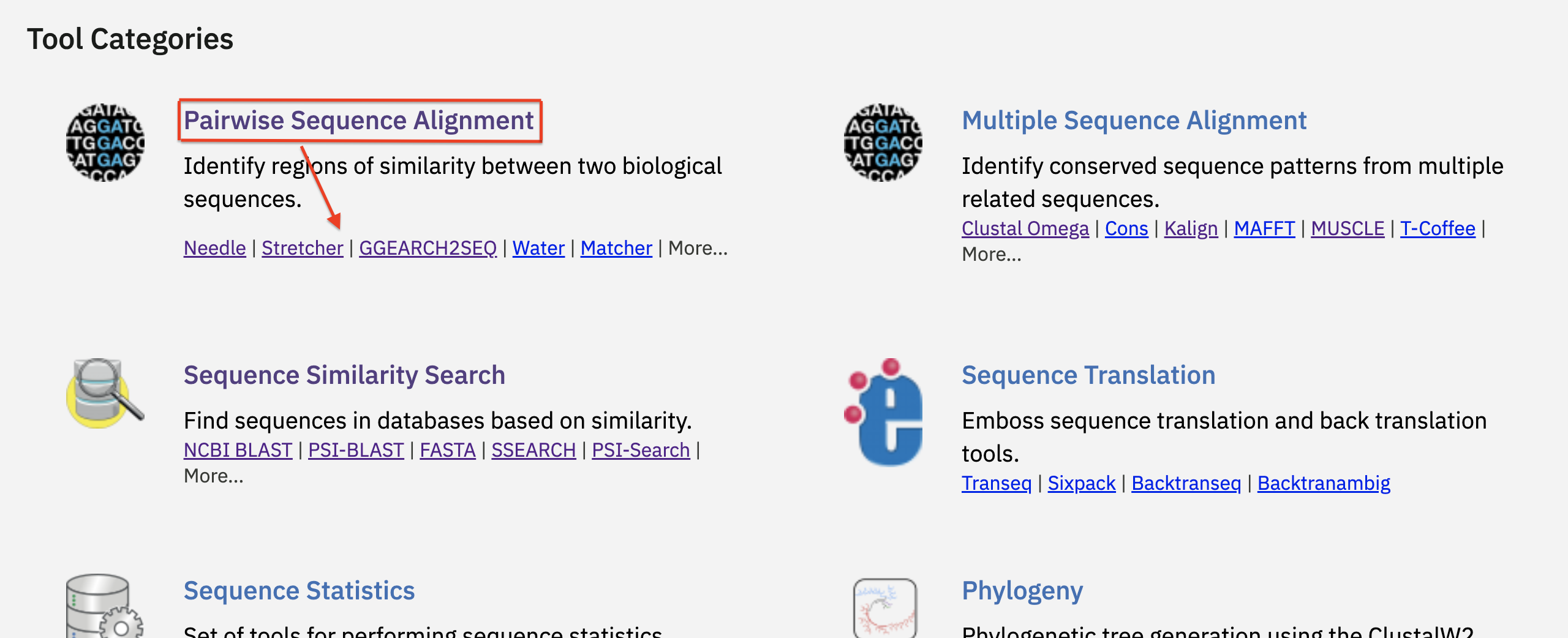
The ‘Tool Category’ section assists in identifying the appropriate tool for your analysis. There are 12 tool categories. Each category briefly describes what it does and lists tools in that category. Links are provided here.
The front page also has a section for the latest news and Twitter updates. We have also listed a few of our collaborators.
Tool webforms
Running a tool from the web form is a simple multiple steps process. Each tool has at least 2 steps, but most of them have more, based on the tool category:
- The first steps are usually where the user sets the tool input (e.g. provide your input protein/nucleotide sequences, specify the databases (in case of sequence similarity search). The input sequence should be in one of the tool-specified formats. You can either paste the sequence or use the file upload option.
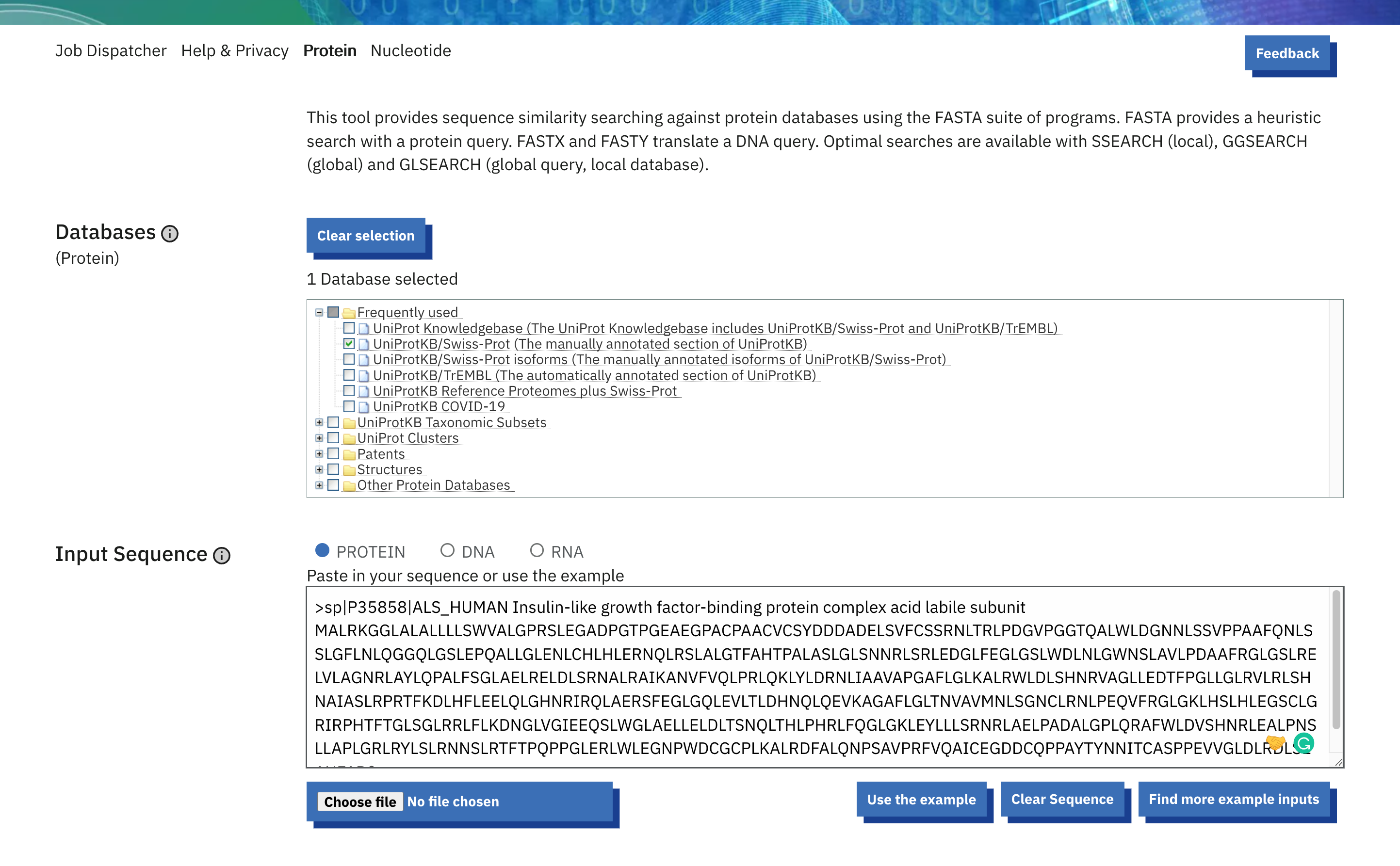
- In the following steps, the user has the possibility to change the default tool parameters. Click on ‘More Options’ to see all the parameters for that specific tool. Mouse over the parameters to get more information about the parameter and default values.
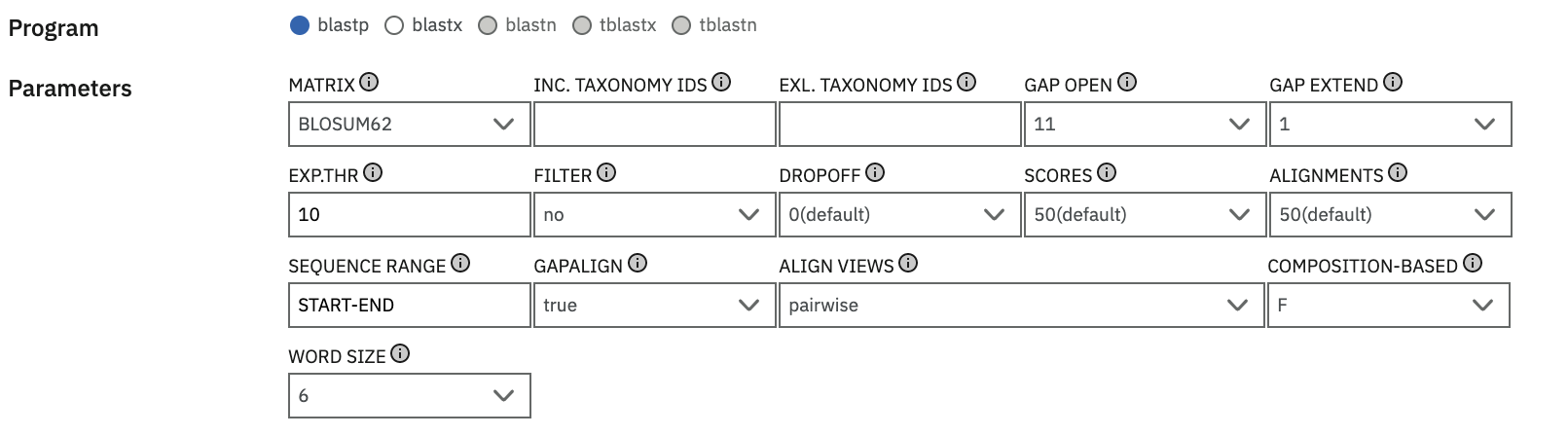
- And finally, the last step is always the tool submission step. Using the submit button will effectively submit the information specified previously in the form to launch the tool on the server.
Input validation
When a job is submitted, the webform performs some basic validation, for example, to check if any input has been provided. Additional validation is performed on the backend server to fully validate the input.
Tip
Parameters are validated by the backend server prior to running the tool on our HPC and in the event of a missing or wrong combination of parameters, the user will be notified directly in the webform.
In addition to input validation, note that there are input constraints that vary across different tools. These can be on the number of input sequences, file size, in the case that the input is provided as a file upload, as well as the parameter values. These usually depend on the context of other parameters, the input provided or the program or database selected. The server validation messages typically provide a good hint on what the issue might be.
Warning
There is a file size limit for each tool. If your input sequence exceeds the limit, you will need to download the tool and run it locally.
Sequence databases
Job Dispatcher provides more than 45,000 distinct sequence libraries from major database resources hosted at EMBL-EBI, including UniProtKB, ENA and Ensembl Genomes. These are available for search through sequence similarity search applications.
A complete list of databases provided by Job Dispatcher is provided in the next section Available databases.
Dbfetch
Dbfetch provides a common interface to database entry retrieval in a variety of different formats.
Dbfetch provides all the sequence libraries available to search in Job Dispatcher, with addition of several metadata datasets, including EMDB, PDBe-KB, MEDLINE, NCBI Taxonomy, EDAM ontology and HGNC.
A list of databases available in Dbfetch is provided on the Dbfetch Databases page.